Researchers in the United States and Japan have provided novel insights into the pathologic mechanisms underlying coronavirus disease 2019 (COVID-19) that may aid the identification of potential diagnostic biomarkers and treatment targets.
The team conducted a gene co-expression network analysis that identified three important gene clusters or “modules” that were closely related to COVID-19.
The study revealed that the three key modules were mostly associated with platelet secretion and activation pathways, mitochondrial processes, and apoptosis, pointing to the hyperactivation of platelets in COVID-19.
“Collectively, our results offer a new and interesting insight into platelet involvement in COVID-19 at the molecular level, which might aid in defining new targets for the treatment of COVID-19–induced thrombosis,” says the team from Texas A&M University in Kingsville and the National Institutes of Biomedical Innovation, Health and Nutrition in Osaka.
A pre-print version of the research paper is available on the bioRxiv server, while the article undergoes peer review.
COVID-19-induced thrombosis increases the risk of cardiovascular occlusive events
The COVID-19 pandemic has impacted tens of millions of people worldwide, and in the United States alone, more than 40 million cases and more than 650,000 deaths have been reported since the outbreak began in late December 2019.
Several studies have now shown that COVID-19-induced thrombosis increases the risk of cardiovascular occlusive events among patients. In addition, researchers have also established that COVID-19 increases platelet activation, which is a primary driver of this thrombotic outcome.
“However, the comprehensive molecular basis of platelet activation in COVID-19 remains elusive,” says Fadi Khasawneh and colleagues. “While there has been some progress, our understanding of the pathways that govern platelet participation in COVID-19-induced thrombosis remains limited, but clearly warrants investigation.”
Where does co-expression analysis come in?
Several computational and research methods have been developed to help provide comprehensive insights into the pathological mechanisms underlying specific disease states.
Some of these methods have been used to examine the potential gene networks involved and are highly instrumental in guiding the understanding of diseases and their mechanistic pathways.
One such approach is co-expression analysis, which clusters genes into co-expressed groups known as modules.
Genes belonging to the same module are thought to share functional properties, meaning co-expression analysis enables researchers to identify associations between gene products and phenotypes.
“In fact, co-expression using weighted correlation network analysis (WGCNA) has been used for analyzing a number of biological processes, including cancer and cognitive and mental disorders,” says Khasawneh and the team.
“Previous studies on the mechanisms of thrombosis in COVID-19 have primarily concentrated on specific pathophysiological functions, with relatively fewer studies identifying comprehensive regulatory networks,” they add.
What did the researchers do?
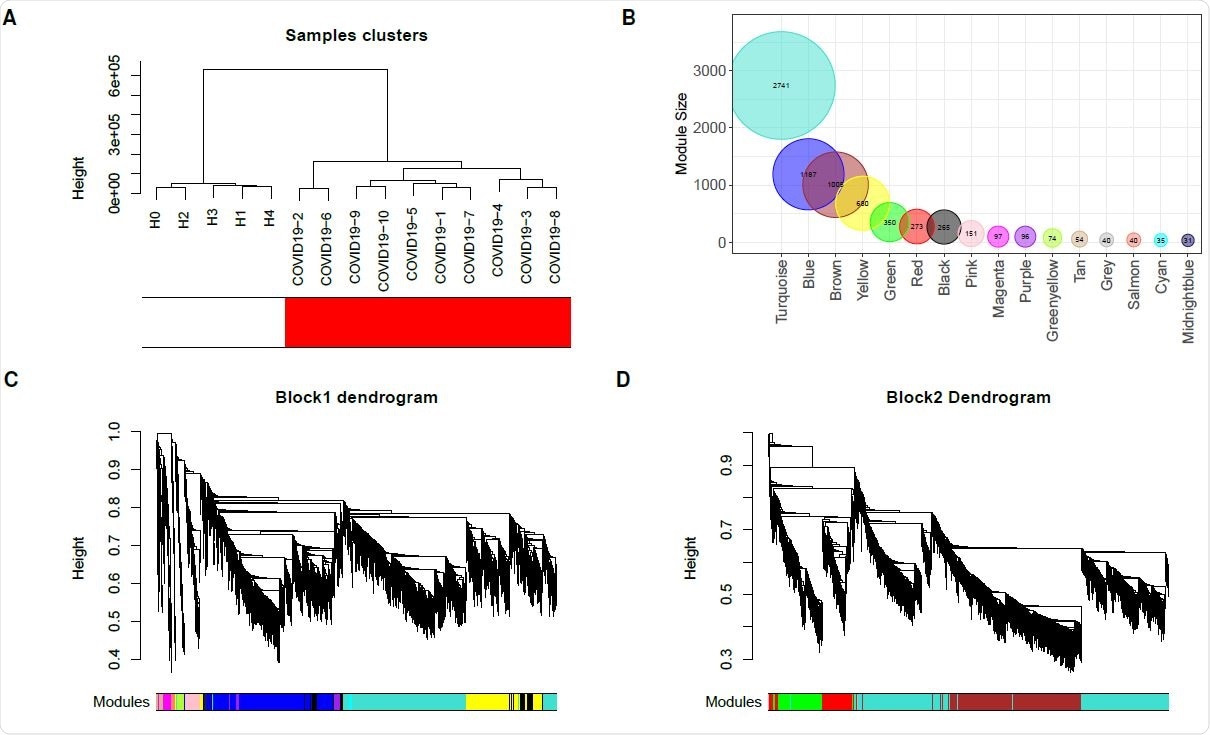
The researchers performed WGCNA on a platelet RNA sequencing dataset containing gene expression data from ten COVID-19 patients and five healthy controls.
Three modules exhibiting the most significant correlation with COVID-19 were identified.
Gene enrichment analysis showed that these three modules were mainly associated with platelet metabolism, protein translation, mitochondrial activity, oxidative phosphorylation, and apoptosis, pointing to a hyperactive state of the platelets.
The top three genes within each of these key modules with the highest intramodular connectivity were selected as the hub genes for COVID-19. These genes included COPE, CDC37, CAPNS1, AURKAIP1, LAMTOR2, GABARAP, MT-ND1, MT-ND5, and MTRNR2L12.
What are the implications of the study?
The researchers say the findings at least partly illustrate the comprehensive platelet regulatory network involved in COVID-19.
“These data should help expand our understanding of the underlying mechanisms of thrombosis in COVID-19 disease and help promote and guide future experimental studies to investigate the roles of the protein-coding genes in the pathophysiology of this disease,” says Khasawneh and colleagues.
“Additionally, these genes may serve as novel therapeutic targets for treating patients,” they conclude.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.